소량의 세포 (10,000개의 세포)로부터 ChIP-seq 분석 기법 확립
- 데이터 제공: Prof. Shirahige Katsuhiko
(The Institute for Quantitative Biosciences (IQB), The University of Tokyo)
◆ Introduction
크로마틴면역침강법 (ChIP; chromatin immuno precipitation)은 목적 단백질이 결합되어 있는 염색체 영역을 면역침강법으로 농축 (enrichment)하는 기술이다. Chromatin Immunoprecipitation Sequencing (ChIP-seq)은
농축한 영역을 Next Generation Sequencing (NGS)으로 분석하는 방법으로, 에피제네틱스 연구를 위한 필수적인 기술이다. ChIP-seq은 사용하는 세포의 수가 적고 ChIP으로부터 회수되는 DNA의 양이 매우 적어,
미량의 ChIP DNA 분석법을 확립하는 것은 매우 중요한 과제이다. 이에 따라, 본 연구자는 크로마틴 단편화 (Chromatin fragmentation)의 최적화 및 다카라바이오의 SMARTer
® ThruPLEX
® DNA-seq Kit을 이용하여,
미량의 ChIP DNA-seq 분석 기법을 확립하였다.
◆ 사용제품
SMARTer® ThruPLEX® DNA-seq Kit (Takara)
◆ Workflow
 ◆ NGS Mapping 결과
◆ NGS Mapping 결과
1x10
4개 또는 2.5×10
5개의 망막색소상피세포 (RPE)에서 H3K4me3 항체를 이용해 ChIP DNA 샘플을 얻었다. 면역 침강하지 않은 세포를 “Input” 샘플로 표기하였다. ChIP DNA 샘플의 전량을
이용해 SMARTer
® ThruPLEX
® DNA-seq Kit로 NGS Library를 제작하여 염기서열을 분석하였다. 1x10
4개의 적은 세포 샘플에서 duplicate rate가 9.1%로 확인되었고 2.5×10
5 개
세포 샘플의 실험 결과와 유사하였다.
샘플 |
Total reads |
Mapped reads |
Unmapped reads |
PCR Bias
(duplicate) |
Input |
31,400,938 |
26,696,740 (78.7%) |
3,087,220 (9.8%) |
3,616,978 (11.5%) |
1x104 세포
(H3K4me3) |
30,823,404 |
21,336,878 (69.2%) |
6,697,451 (21.7%) |
2,789,075 (9.1%) |
2.5x105 세포
(H3K4me3) |
45,738,702 |
37,730,704 (82.5%) |
4,018,338 (8.8%) |
3,989,660 (8.7%) |
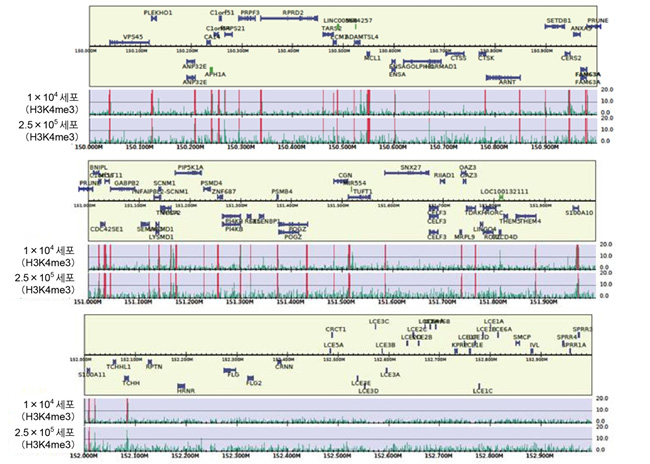
◆ Peak 검출결과
1×10
4 세포로부터 얻은 H3K4me3의 peak profile은 2.5×10
5 세포의 결과와 유사함을 확인하였다.
◆ 제품 사용을 검토 중인 분께 사용자의 한마디
SMARTer® ThruPLEX® DNA-seq Kit을 이용하면 소량의 DNA로도 ChIP-seq 분석을 할 수 있습니다. 물론, 샘플량이 소량인지 아닌지를 확인하는 것이 어려운 분에게도 매우 추천합니다.
간편하고 효율적으로 ChIP DNA의 NGS Library를 제작할 수 있기 때문에, 처음 실험하는 분께도 매우 추천할 만한 제품입니다. (Prof. Shirahige Katsuhiko, The University of Tokyo)





